The evolution of contact prediction: my new paper
I'm so pleased to be able to write about my work, with Saulo de Oliveira, Konrad Krawczyk, and Charlotte Deane, on our paper "The evolution of contact prediction: evidence that contact selection in statistical contact prediction is changing" (Bioinformatics btz816). Contact prediction - the prediction of parts of the amino-acid chain that are close together - has been critical to improving the ability of scientists to predict protein structures over the last decade. Here we look at the properties of these predictions, and what that might mean for their use.
The paper begins with a question. If contact prediction methods are based on statistical properties of sequence alignments, and those alignments are generated in the presence of ecological and physical constraints, what effect do the physical constraints have on the statistical properties of real sequence alignments? More concisely: when we predict contacts, do we predict particularly important contacts?
We used statistical models and machine-learning predicted contacts for more than 800 protein domains with structures which had different evolutionary histories. By comparing the predicted contacts to the contacts which were not predicted, we were able to comment on the types of predictions that the methods made.
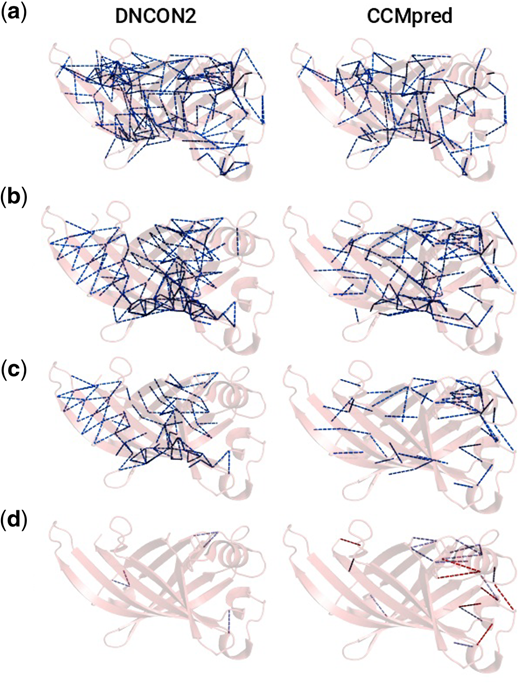
Here's an example. DNCON2 is a deep-learning-based contact predictor from Badri Adhikari and coauthors, while CCMpred (from the Söding group) implements a Potts model scheme, which directly measures coevolutionary pressures between residues which have been recorded in the alignment. We would expect these pressures to be greater for 'more important' contacts, but that deep learning might lose some of this signal in favour of contacts which are in some ways 'easier to predict'.
In A, we see a randomly-selected set of contacts that were not predicted. There are fewer for CCMpred because we only selected as many as were correctly predicted among the top L predictions. B shows the distribution of correct predictions among the top L predictions for each method, and here we see differences. DNCON2 tends to 'lace up' the beta sheets, while CCMpred heavily predicts contacts between the upper-right alpha helix and the beta barrel. The differences are magnified in C, which shows only those contacts which are predicted either by CCMpred or DNCON2 but not both. D shows a striking difference between the distributions of correctly-predicted contacts associated with chemical bonds that are not within a secondary structure, suggesting that CCMpred may reveal contacts that are biophysically more interesting.
There's plenty more in the paper. Give it a read and let us know what you think.